CecogAnalyzer
by
held
—
last modified
Jul 24, 2015 10:51 AM
![]()
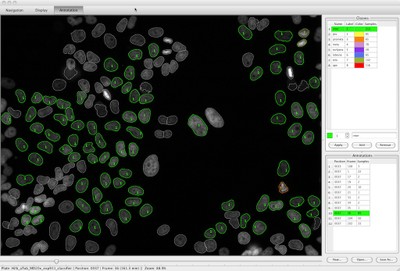
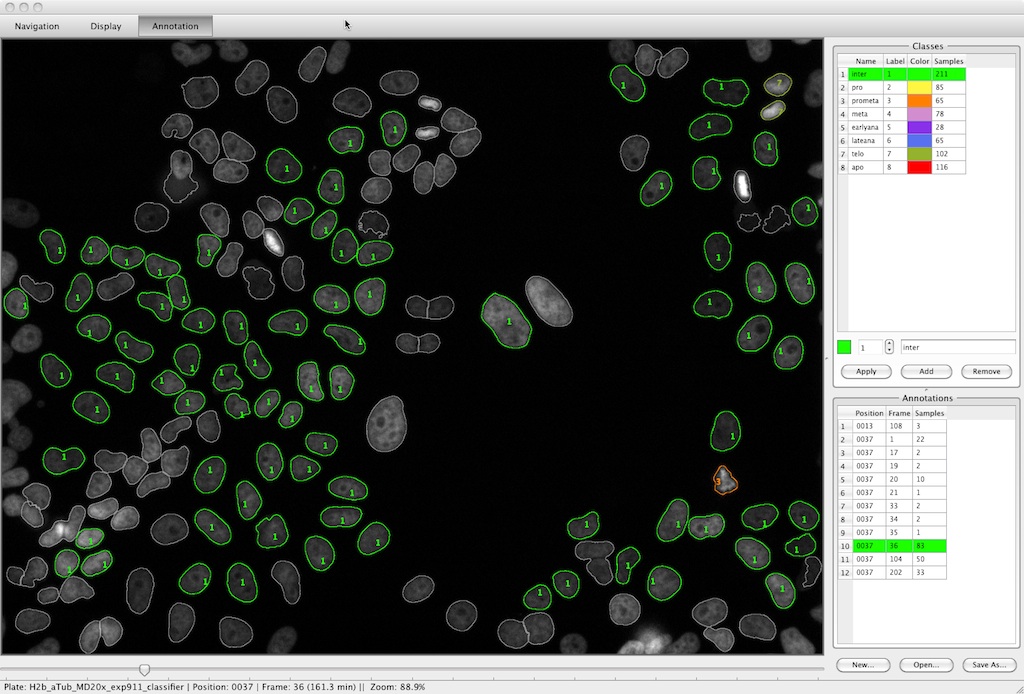
The CecogAnalyzer is a stand-alone application built on top of the CellCognition framework. A graphical user interface provides a comfortable setup and parameterization of the analysis workflow.
Features are:
- Object detection
- Feature extraction
- Classification
- Tracking of individual cells over time
- Detection of class-transition motifs (e.g. cells entering mitosis)
- Correction of classification errors on the detected event tracks
- The workflow can either be executed either on the local computer or as a batch process on any cluster environment.
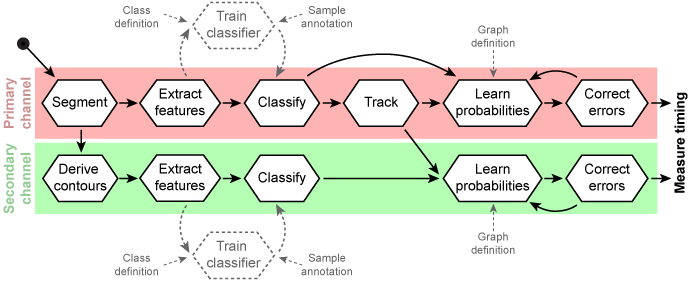
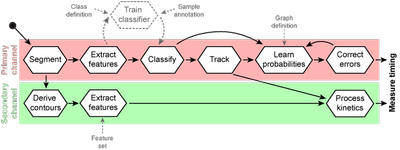
Generic workflow for automated annotation of cellular dynamics
Based on the primary channel objects were detected and contours derived for the secondary channel. Features were extracted from both channels independently. An additional step of classifier training is necessary if no suitable classifier is present. This step must by applied once for the given marker and imaging conditions. Object labels are predicted with probability estimates and primary objects are tracked over time. Secondary are linked to primary objects and therefore tracked indirectly. Tracks are selected by stage transition motifs and in silico aligned. The error correction by hidden Markov model is iteratively coupled with the EM-algorithm for optimization. Despite the derived contours are all steps executed independently for the primary and secondary channel in order to correlate timing differences later. For classifier training information about classes and samples are necessary. A human expert annotates image objects into one of the predefined classes. Transition probabilities for error correction are learned by default on a fully connected graph. To constrain stage transitions a user-defined graph structure can be applied.
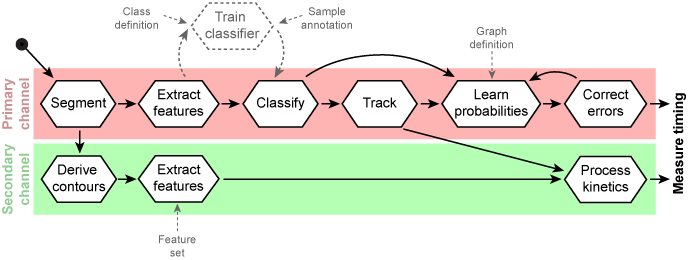
Generic workflow for automated measuring of protein kinetics
Modified version of the workflow above in a way that classification and error correction was removed for the secondary channel. Instead, feature values are recorded along the trajectories and processed in to measure timings, e.g. mitotic exit timing, or to derive protein degradation kinetics, e.g. Securin levels.